| 3X+1@home
|
|
Image:Px.gif
|
|
Image:Px.gif
|
The 3x+1 conjecture is an unsolved conjecture in mathematics. It is also known as the Collatz conjecture (after Lothar Collatz, who first proposed it in 1937), as the Ulam conjecture (after Stanislaw Ulam), as the Syracuse problem, as the hailstone sequence or hailstone numbers, or as wondrous numbers per Gödel, Escher, Bach. It asks whether a certain kind of number sequence always ends in the same way, regardless of the starting number. Paul Erdõs said of the 3x+1 conjecture, "Mathematics is not yet ready for such problems." This project seeks to find large solutions to this conjecture.
Image:Enter arrow.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
| ABC@home
|
|
Image:Px.gif
|
|
Image:Px.gif
|
Help solve one of the greatest open problems in mathematics, the ABC conjecture! The conjecture is stated in terms of simple properties of three integers, one of which is the sum of the other two (a + b = c): 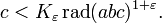 The ABC conjecture has a large number of interesting consequences, including its relationship to questions in number theory, and therefore Fermat's Last Theorem. The ABC conjecture also gives a conditional proof for other conjectures (if the ABC conjecture were true, these other conjectures would be as well). "The ABC conjecture is the most important unsolved problem in Diophantine analysis... It is more than utilitarian; to mathematicians it is also a thing of beauty. Seeing so many Diophantine problems unexpectedly encapsulated into a single equation drives home the feeling that all the sub disciplines of mathematics are aspects of a single underlying unity, and that at its heart lie pure language and simple expressibility." (Goldfeld, Dorian. 1996. "Beyond the last theorem". Math Horizons. September:26-34.) The ABC conjecture has a large number of interesting consequences, including its relationship to questions in number theory, and therefore Fermat's Last Theorem. The ABC conjecture also gives a conditional proof for other conjectures (if the ABC conjecture were true, these other conjectures would be as well). "The ABC conjecture is the most important unsolved problem in Diophantine analysis... It is more than utilitarian; to mathematicians it is also a thing of beauty. Seeing so many Diophantine problems unexpectedly encapsulated into a single equation drives home the feeling that all the sub disciplines of mathematics are aspects of a single underlying unity, and that at its heart lie pure language and simple expressibility." (Goldfeld, Dorian. 1996. "Beyond the last theorem". Math Horizons. September:26-34.)
Image:Enter arrow.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
| Africa@home
|
|
Image:Px.gif
|
|
Image:Px.gif
|
The goal of Africa@home is to use volunteer distributed computing to provide supercomputing resources to African universities and institutions. The first such project is MalariaControl.net, a project to determine optimal strategies for controlling the spread of Malaria. Computer models of the transmission dynamics and health effects of the disease can be used to improve planning for delivery of mosquito netting, medicines, and other resources. But such models would literally take forty years to run on the computers available to the scientists who developed them; with the help of volunteers like yourself we hope get the first results in a few months.
Image:Enter arrow.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
| APS@home
|
|
Image:Px.gif
|
|
Image:Px.gif
|
APS@Home studies problems in atmospheric science. At the moment, we're concentrating on atmospheric dispersion, and how it affects the accuracy of estimates used in global climate models. In the current application, your computer simulates the trajectories of tracer "particles" within and above forest and crop canopies, to improve our understanding of ecosystem - atmosphere interactions. Many climate models depend on details of these interactions; the understanding we gain from the work your computers do helps to improve the quality of such models.
Image:Enter arrow.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
| BBC Climate Change
|
|
Image:Px.gif
|
|
Image:Px.gif
|
The same model that the Met Office uses to make daily weather forecasts has been adapted to run on home PCs. The model incorporates many variable parameters, allowing thousands of sets of conditions. Your computer will run one individual set of conditions-- in effect your individual version of how the world's climate works-- and then report back to the research team what it calculates. This experiment was described on the BBC television documentary Meltdown (BBC-4, February 20th, 2006). Note: workunits require several months of screensaver time; faster computers recommended.
Image:Enter arrow.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
| BOINC Alpha Test
|
|
Image:Px.gif
|
|
Image:Px.gif
|
This is the official test project for BOINC (the software platform underlying GridRepublic). New features and updates will be tested here before being used for production. The goal for of this project to improve BOINC and therefore all BOINC-based projects and services.
Image:Enter arrow.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
| BRaTS@home
|
|
Image:Px.gif
|
|
Image:Px.gif
|
BRaTS@Home is a research project that uses Internet-connected computers to do calculations in Gravitational Ray Tracing, also known as Gravitational Lensing. (BRaTS stands for BRaTS Ray Trace Simulator.) Gravitational Lensing is one of the most useful tools at the disposal of astrophysicists studying dark matter in galaxies and galaxy clusters: Through the study of gravitational deflection of the light of background objects due to foreground objects and structures one can detect the unseen matter which is currently thought to comprise most of the mass of the universe. Brats@home calculations will be used to further study the commonly accepted model of mass and density of the universe (the Navarro, Frenk and White model).
Image:Enter arrow.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
| BURP
|
|
Image:Px.gif
|
|
Image:Px.gif
|
BURP (Big and Ugly Rendering Project) is a public distributed system for rendering 3D animations. Support the arts: Lend your idle computing time to the BURP network, and/or Be an artist: submit (non-commercial) animations for rendering on this network. (Information on submitting files for rendering is here.) Note: This project is still in development, which means that certain restrictions apply. Not all uploaded sessions will actually be rendered right away - and sometimes you will not be able to contact the schedulers for short periods of time.
Image:Enter arrow.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
| Cels@home
|
|
Image:Px.gif
|
|
Image:Px.gif
|
Cels@Home is a research project that uses Internet-connected computers to do research in cell adhesion. One of the many applications of this is in cancer research, as the point at which cancerous cells quit staying in place, and instead break free to move throughout the body, is a critical event that makes the disease much harder to treat.
Image:Enter arrow.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
| Chess960@home
|
|
Image:Px.gif
|
|
Image:Px.gif
|
Chess960 is an innovative and increasingly popular variant of classical Chess. In Chess960, just before the start of every game, the initial configuration of the pieces is determined randomly: the king, queen, rooks, bishops and knights are not necessarily placed on the same home squares as in classical Chess. The goal of Chess960@home is to develop basic game theory for Chess960, such as exists for classical Chess. Due to the many possible variations in starting conditions, establishing such theory for Chess960 is computationally intensive and so we turn to volunteer enthusiasts like you.
Image:Enter arrow.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
| Climateprediction.net
|
|
Image:Px.gif
|
|
Image:Px.gif
|
Climateprediction.net is computing a massive environmental model intended to forecast climate conditions in the 21st century. Climate change, and our response to it, are issues of global importance, affecting food production, water resources, ecosystems, energy demand, insurance costs and much else. There is broad scientific consensus that the Earth will probably warm over the coming century; climateprediction.net should, for the first time, tell us what is most likely to happen.
Image:Enter arrow.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
| Cosmology@home
|
|
Image:Px.gif
|
|
Image:Px.gif
|
A Universe in your PC: Cosmology@home seeks to learn how the physical universe took it's shape. Towards that end, the project is distributing large numbers of "simulation packages", each a simulated universe with assumptions about the geometry of space, particle content, and "physics of the beginning". Results of these simulations will be compared to real-world observations, in order to see which (if any) of the initial assumptions best match the world in which we live. In order to produce meaningful results the project needs to run millions of these simulations (or to get stupendously lucky), so your help is essential.
Image:Enter arrow.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
| DepSpid
|
|
Image:Px.gif
|
|
Image:Px.gif
|
DepSpid is a distributed web crawler, much like those used by search engines to index the Internet-- except that DepSpid is powered by volunteer PCs rather than a corporate data center. The major goals of DepSpid are (1) to collect statistical data about the structure of the web, and (2) to build a database containing the dependencies between individual web sites and groups of web sites. All information collected by the spider will be made publicly available. Note: DepSpid uses your computer and Internet connection to analyze random web sites. The owner of the Internet connection needs to know that the computers running DepSpid may download HTML pages of sites which you would never visit yourself. DepSpid will download HTML pages only and never other things like executables, images or other elements which may contain dangerous content. However, in some countries (and at some companies) it may be forbidden to access specific sites or kinds of sites. If you are subject so such restrictions, you should not run DepSpid.
Image:Enter arrow.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
| Docking@home
|
|
Image:Px.gif
|
|
Image:Px.gif
|
Docking@Home is a collaborative project that aims to accomplish both bioscience and computer science goals. From the bioscience point of view, the project aims to further knowledge of the atomic details of protein-ligand interactions and, by doing so, to facilitate the discovery of novel pharmaceuticals. From the computer science point of view, the project aims to extend volunteer computing to enable adaptive multi-scale modeling: different models that represent the same phenomena in nature with different levels of accuracy and resource requirements will be chosen at run-time based on results collected previously and on characteristics of the protein-ligand complex being studied.
Image:Enter arrow.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
| Einstein@home
|
|
Image:Px.gif
|
|
Image:Px.gif
|
According to Albert Einstein, we live in a universe full of gravitational waves. He suggested that the movements of heavy objects, such as black holes and dense stars, create waves that change space and time. We have a chance to detect these waves, but we need your help to do it! With Einstein@home, you can contribute your computer's idle time to a search for spinning neutron stars (also called pulsars) using data from the LIGO and GEO gravitational wave detectors. Einstein@home is a World Year of Physics 2005 project supported by the American Physical Society (APS) and by a number of international organizations.
Image:Enter arrow.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
| Enigma@home/M4
|
|
Image:Px.gif
|
|
Image:Px.gif
|
The "Enigma" was a cipher machine used to encrypt and decrypt secret messages. The device was used commercially from the early 1920s on, and was also adopted by the military and governmental services of a number of nations--most famously by Nazi Germany before and during World War II. This project seeks to break 3 original Enigma messages with the help of distributed computing. The signals were intercepted in the North Atlantic in 1942 and are believed to be unbroken. Ralph Erskine has presented the intercepts in a letter to the journal Cryptologia. The signals were presumably enciphered with the four rotor Enigma M4 - hence the name of the project.
Image:Enter arrow.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
| Hydrogen@home
|
|
Image:Px.gif
|
|
Image:Px.gif
|
Tired of melting polar ice caps? Upset by nations fighting over oil? Join Hydrogen@home and help develop an alternative energy technology that is clean, abundant, and locally controlled. The US DOE has engineered algae that convert light energy to hydrogen (info). This hydrogen can be used to power cars, buses, houses and other things; when burned it produces no pollution, only water. But: the process is not yet sufficiently efficient for large scale use. With the help of volunteers like you, Hydrogen@Home is modeling enzymes that could significantly improve the effectiveness of the biochemical processes involved. So please join us-- with your help, we may soon be able to grow 100% clean fuel instead of drilling or mining for fossil fuels.
Image:Enter arrow.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
| Lattice Project
|
|
Image:Px.gif
|
|
Image:Px.gif
|
The Lattice Project supplies computing power to scientists at the University of Maryland. The volunteered resources are integrated with on-campus Grid Resources, and so no single project or application dominates the landscape. Some projects include: Study of evolutionary relationships based on DNA sequence data; Analysis of bacterial, plasmid, and virus protein sequences; And study of biological diversity in nature reserves. For more information about currently-running research, click here.
Image:Enter arrow.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
| Leiden Classical
|
|
Image:Px.gif
|
|
Image:Px.gif
|
Classical Mechanics is the physics used to describe the motion of objects in the observable universe-- everything from baseballs to bullets, from bicycles to spacecraft, from raindrops to planets, stars, and galaxies. Leiden Classical-- with your help-- provides a computing service which allows scientists and students (and the general public) to submit particle-system simulation jobs; the project is thus a resource for both research and education. To create a dataset and submit it for simulation, click here.
Image:Enter arrow.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
| LHC@home
|
|
Image:Px.gif
|
|
Image:Px.gif
|
CERN (European Center for Nuclear Research) is currently constructing the Large Hadron Collider (LHC), the world's largest and most powerful particle accelerator. LHC@home helps construction of the LHC by simulating how particles travel through the 27 km long accelerator tunnel. With the help of these calculations, the magnets that control the particle beam can be calibrated with greater precision, making it possible to smash together individual atoms at nearly the speed of light. By observing the aftermath of these collisions, physicists hope to learn what matter is made of and what forces hold it together.
Image:Enter arrow.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
| Milkyway@home
|
|
Image:Px.gif
|
|
Image:Px.gif
|
MilkyWay@home is analyzing data from the Sloan Sky Survey, with the goal of discovering structures existing in the Milky Way galaxy and their spatial distribution. By identifying and quantifying the structure of the Milky Way, it will be possible to test models for the formation of our galaxy, and by example the process of galaxy formation in general.
Image:Enter arrow.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
| MindModeling@home
|
|
Image:Px.gif
|
|
Image:Px.gif
|
MindModeling@Home is a research project that uses volunteer computing for the advancement of cognitive science. Our research utilizes and develops rich computer models of cognitive process to better understand the human mind. Current areas of research include: Physical and behavioral impacts of sleep deprivation, sleep restriction, and circadian rhythm: A major goal of this research is generating precise predictions about how cognitive performance will be affected, both as a function of a particular sleep history and at particular points in the circadian cycle. Simulation outputs will be compared to real-world experimental data. (more) Language Modeling: The main idea of "Optimality Theory" is that the observed forms of language arise from the interaction between conflicting constraints. This project uses a computer model of this theory to analyze conversational transcripts. (more) Situated Language and Active Vision: The modeling efforts in this research thread explore how people use spatial language and spatial reasoning skills to communicate and share understanding of situations. (more)
Image:Enter arrow.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
| NanoHive@home
|
|
Image:Px.gif
|
|
Image:Px.gif
|
Nanotechnology is the understanding and manipulation of matter at the molecular scale. At this scale, physical and chemical properties of elements are very different from material at larger sizes. NanoHive@home was founded to help research this important science through simulations and analysis otherwise too complex to be calculated by normal means. NanoHive@home runs projects and simulations that evaluate: (1) nano-scale mechanisms for movement and transport molecules and (2) efficiency of nanobots in combating a potential germ. A broad range of vital industries already benefits from advances made in nanotechnology. In the field of medicine, therapeutics, cancer treatments, and nerve and tissue repair all look to nanotechnology. In computer sciences, nanotechnology is already being used to bring circuitry to the molecular level, increasing processing power and speed.
Image:Enter arrow.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
| NQueens@home
|
|
Image:Px.gif
|
|
Image:Px.gif
|
This project uses Internet-connected computers to solve the "N Queens" puzzle: the problem of putting eight chess queens on an 8x8 chessboard such that none of them is able to capture any other using the standard chess queen's moves. The queens must be placed in such a way that no two queens would be able to attack each other. Thus, a solution requires that no two queens share the same row, column, or diagonal.
Image:Enter arrow.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
| Pirates@home
|
|
Image:Px.gif
|
|
Image:Px.gif
|
AAARGH! Welcome, mates, to Pirates@Home! Pirates@Home is an ongoing test of BOINC, the Berkeley Open Infrastructure for Network Computing. Pirates@Home is not doing any real scientific computation. Our goal is to provide a testbed for new projects and features. (BOINC is the software platform used by GridRepublic.) Please note that Pirates@Home is always under development; please don't expect it to work all the time.
Image:Enter arrow.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
| POEM@home
|
|
Image:Px.gif
|
|
Image:Px.gif
|
Proteins are the machinery of all known cellular life. Amazingly, these large biomolecules with up to 100,000 atoms fold into unique three-dimensional shapes; these shapes determine each protein's function. But the shape of a protein is very difficult to determine experimentally. So POEM@HOME implements a novel approach to determining protein structure computationally. (The scientific approach behind POEM@HOME is based on the thermodynamic hypothesis that won C. B. Anfinsen the Nobel Prize in Chemistry in 1972.) By joining this project you will help to
- predict the biologically active structure of proteins
- understand the signal-processing mechanisms by which proteins interact with one another
- understand diseases related to protein malfunction or aggregation
- develop new drugs on the basis of the three-dimensions structure of biologically important proteins
Image:Enter arrow.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
| Predictor@home
|
|
Image:Px.gif
|
|
Image:Px.gif
|
Predictor@home seeks to address critical biomedical questions of protein related diseases such as Cancer. Specifically, P@H uses distributed computing resources to test and evaluate new algorithms and methods for "protein folding": that is, new ways of predicting protein structure from amino acid sequence. Improved understanding of the relationship between protein structure and sequence will open new avenues of inquiry and discovery in biology and medicine, by making visible for the first time the basic mechanics of many essential life functions.
Image:Enter arrow.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
| PrimeGrid
|
|
Image:Px.gif
|
|
Image:Px.gif
|
Currently, PrimeGrid has two projects: (i) PrimeGen, which is working to build a public database of prime numbers, and (ii) Twin Prime Search, which searches for pairs of prime numbers which are separated by two- for example 5 and 7, 11 and 13, 821 and 823. Prime Numbers are of great interest to mathematicians for a variety of reasons. Primes also play a central role in the cryptographic systems which are used for computer security. Through the study of Prime Numbers it can be shown how much processing is required to crack an encryption code and thus to determine whether current security schemes are sufficiently secure.
Image:Enter arrow.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
| Project Neuron
|
|
Image:Px.gif
|
|
Image:Px.gif
|
The subject of this project is volunteer computing itself. Specifically, the goal is to develop metrics that will be useful to establish or refute the quality/reliability/dependability of this and other volunteer computing projects.
Image:Enter arrow.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
| Proteins@Home
|
|
Image:Px.gif
|
|
Image:Px.gif
|
Proteins@Home is investigating the "Inverse Protein Folding Problem": Whereas "Protein Folding" seeks to determine a protein's shape from its amino acid sequence, "Inverse Protein Folding" begins with a protein of known shape and seeks to "work backwards" to determine the amino acid sequence from which it is generated. Inverse Protein Folding has the potential to significantly impact future drug design by providing tools to develop novel proteins with specific structural (and functional) properties.
Image:Enter arrow.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
| PS3Grid
|
|
Image:Px.gif
|
|
Image:Px.gif
|
NOTE: THIS PROJECT WILL RUN ONLY ON PS3 CONSOLES IF YOU DO NOT HAVE A PS3, IT WILL NOT WORK. PS3GRID is a volunteer computing project on the PlayStation3 using Molecular Dynamics (MD), a methodology for simulating the interactions of atoms and molecules over time. By applying known laws of physics to the simulation environment and objects, MD enables, in effect, "virtual experiments". MD is used in academia and pharma companies for a wide range of applications including drug design, drug screening, and in general to investigate protein function. A single PS3 can provide processing power equal to about 20 single-core computers; 1000 PS3s will be the equivalent of 20,000 computers. Installation can be done in five easy steps with a simple USB drive. (more information and help) NOTE: THIS PROJECT WILL RUN ONLY ON PS3 CONSOLES IF YOU DO NOT HAVE A PS3, IT WILL NOT WORK.
Image:Enter arrow.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
| Quantum Monte Carlo
|
|
Image:Px.gif
|
|
Image:Px.gif
|
Reactions between molecules are important for virtually all parts of our lives. The structure and reactivity of molecules can be predicted by Quantum Chemistry, but the solution of the vastly complex equations of Quantum Theory often require huge amounts of computing power. This project seeks to raise the necessary computing time in order to further develop the very promising Quantum Monte Carlo (QMC) method for general use in Quantum Chemistry.
Image:Enter arrow.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
| Rectilinear Crossing
|
|
Image:Px.gif
|
|
Image:Px.gif
|
The Rectilinear Crossing Number Project examines a basic problem of geometry, with application in a range of practical problems from transportation to printing. Imagine a finite set of points on a flat surface. Draw a graph, such that each point is connected to every other point with a straight line. Now count: how many times do the lines cross? The "Rectilinear Crossing Number" is the minimum number of crossings for that set of points, realized when the points are optimally arranged. The challenge is that as the number of points increases, determining this optimal arrangement becomes exceptionally difficult. The main goal of the current project is to determine the "Rectilinear Crossing Number" for 18 points, which is as yet undetermined.
Image:Enter arrow.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
| Riesel Sieve
|
|
Image:Px.gif
|
|
Image:Px.gif
|
Riesel Sieve is a distributed effort to solve the "Riesel problem": Riesel's Theorem hypothesizes that there exist infinitely many odd integers k such that k*2n - 1 is composite for every n > 1. Riesel showed that k = 509203 has this property, and also the multipliers kr = k0 + 11184810r for r = 1, 2, 3, . . . Such numbers are now called Riesel numbers because of their similarity with the Sierpinski numbers. The "Riesel problem" consists in determining the smallest Riesel number. The Riesel Sieve project uses volunteered computing resources to remove prime candidates from the roughly 11 million possibilities, thus gradually closing in on potential solutions.
Image:Enter arrow.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
| Rosetta@home
|
|
Image:Px.gif
|
|
Image:Px.gif
|
For every disease there is a cure: you. Rosetta@home needs your help to determine the 3-dimensional shape of proteins as part of research that may ultimately contribute to cures for major human diseases such as AIDS / HIV, Malaria, Cancer, and Alzheimer's. By running the Rosetta program on your computer while you're not using it, you will help us speed up and extend our research in ways we couldn't possibly attempt without your help.
Image:Enter arrow.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
| SciLINC
|
|
Image:Px.gif
|
|
Image:Px.gif
|
Scientific Literature Indexing on Networked Computers (SciLINC) is a public-resource computing application that automatically indexes large amounts of digitized scientific literature, ultimately providing users with an integrated Web portal, available at www.botanicus.org, for the discovery of information about plants. Our project analyzes text from digitized botanical literature in order to return a full-text index and a keyword index for each page. These keywords are annotated with links to other online resources- i.e. Web pages about a particular plant- allowing users of the portal to search for terms, discover where they reside in a body of digitized literature, view the appropriate pages, and click through to discover other online resources associated with that keyword. This Web portal will be an essential tool for anyone interested in learning about plants, including scientists, students, and the general public.
Image:Enter arrow.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
| Seasonal Attribution Project
|
|
Image:Px.gif
|
|
Image:Px.gif
|
Recent extreme weather events have prompted the debate about effects of human activity on the world's climate. Now you can help us to determine the extent to which extreme weather events like the United Kingdom floods of Autumn 2000 are attributable to human-induced climate change. We invite you to download and run high-resolution simulations of the world's climate on your own computer. By comparing the results of these simulations, half of which will include the effects of human-induced climate change, and half of which will not, we will investigate the possible impact of human activity on extreme weather risk. Thank you for your help, and please join the project! NOTE: No new work will be issued for the Seasonal Attribution project from this site. Instead, we will be migrating the project to Climateprediction.net.
Image:Enter arrow.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
| SETI@home
|
|
Image:Px.gif
|
|
Image:Px.gif
|
SETI (Search for Extraterrestrial Intelligence) is a scientific area whose goal is to detect intelligent life outside Earth. One approach, known as radio SETI, uses radio telescopes to listen for narrow-bandwidth radio signals from space. Such signals are not known to occur naturally, so a detection would provide evidence of extraterrestrial technology. Radio telescope signals consist primarily of noise (from celestial sources and the receiver's electronics) and man-made signals such as TV stations, radar, and satellites. Modern radio SETI projects analyze the data digitally. More computing power enables searches to cover greater frequency ranges with more sensitivity. Radio SETI, therefore, has an insatiable appetite for computing power.
Image:Enter arrow.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
| SHA-1 Collision Search Graz
|
|
Image:Px.gif
|
|
Image:Px.gif
|
How secure is secure? The hash function "SHA-1" is one of the most widely-used cryptographic algorithms in the world. It was designed by NSA and put forward as a standard by NIST in 1995. Purchases at online stores, storing and comparing of website passwords, and securing and administering webservers are just a few examples where SHA-1 is used and trusted by many of us, directly and indirectly, on a daily basis. The goal of "SHA-1 Collision Search Graz" is to search for weaknesses (called "collisions") in this essential algorithm. The purpose is not to cause harm, but to verify or disprove the security of this essential building block; and to develop tools which will help to more accurately estimate the security of successors to SHA-1 which are already in development.
Image:Enter arrow.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
| SIMAP
|
|
Image:Px.gif
|
|
Image:Px.gif
|
SIMAP (Similarity Matrix of Proteins) is a public database of protein similarities that plays a key role in bioinformatics research. The SIMAP database contains substantially all currently published protein sequences and is continuously updated. But as the number of sequences grows, so also does the computational effort required to keep the similarities matrix up to date-- the computational effort to calculate the similarity data depends on the square of the number of sequences. Please help to maintain this vital resource by calculating protein similarities on your computer. The computing power you donate supports a wide range of biological research projects that make use of SIMAP data.
Image:Enter arrow.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
| Spinhenge
|
|
Image:Px.gif
|
|
Image:Px.gif
|
Spinhenge is researching the behavior of nano-magnetic molecules (or "single magnetic molecules"). These molecules may be used to develop tiny magnetic switches, with applications ranging from medicine (ex. local tumor chemotherapy), to computer technology (ex. miniature memory modules), to biotechnology.
Image:Enter arrow.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
| Sudoku
|
|
Image:Px.gif
|
|
Image:Px.gif
|
Sudoku is a very popular numbers-based puzzle. If you have played it, you may have noticed that as the number of numbers given decreases, the level of difficulty in finding the solution increases. But, what is the fewest number of numbers that can be given and still have a valid puzzle? How difficult can the puzzle get? Help us find out!
Image:Enter arrow.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
| Superlink@Technion
|
|
Image:Px.gif
|
|
Image:Px.gif
|
Superlink@Technion helps geneticists all over the world find genes causing hereditary diseases, such as some types of diabetes, hypertension (high blood pressure), cancer, schizophrenia and many others. More specifically, Superlink@Technion performs statistical analysis to compare large sets of real genetic data, in order to identify mutated disease-provoking genes. This statistical analysis is called "linkage analysis". Geneticists submit data for the analysis via the Internet, by using the Superlink-online portal. The tasks are then automatically parallelized and invoked on many compute clusters, grids and volunteer computers via Superlink@Technion. This technology has already proved successful for finding various genetic diseases (more info). This method is extremely powerful, but it is also extremely intensive computationally. With your help we can pursue more comprehensive analyses on an unprecedented scale, and hopefully allow for significant advances in the understanding of the mechanisms of disease.
Image:Enter arrow.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
| SZTAKI Desktop Grid
|
|
Image:Px.gif
|
|
Image:Px.gif
|
SZTAKI Desktop Grid searches for generalized binary number systems-- the aim of the project is to find all generalized binary number systems up to dimension 11. It is hoped that the results will have application in data compression, coding, and cryptography, and that the project will lead to a deeper understanding of the mathematics of generalized number systems.
Image:Enter arrow.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
| Tanpaku
|
|
Image:Px.gif
|
|
Image:Px.gif
|
The recently completed Human Genome Project has determined the amino acid sequence of all human proteins, but the function of most these proteins remain unknown. Since there is a deep relationship between the physical shape of a protein and its function in the body, what is needed is a way to determine a protein's shape (unknown) from its amino acid sequence (known). The Tanpaku Project seeks to do exactly that; to predict protein structure and function from known genetic sequences, using the Brownian Dynamics (BD) method, which we believe is much more efficient than other methods.
Image:Enter arrow.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
| TSP
|
|
Image:Px.gif
|
|
Image:Px.gif
|
The Traveling Salesman Problem (TSP) is simple enough: For a given set of cites, how do you identify the shortest route that will allow you to visit visit each city exactly once? This deceptively simple problem is trivial given a small set of cities, however, as you add more cities the number of possible routes goes through the roof. For instance, a set of 48 cities would require evaluation of a staggering number of possible routes: 129,311,620,755,584,000,000,000,000,000,000,000, 000,000,000,000,000,000,000,000 Who cares about traveling salesmen? Do we really need to know how to get a traveling salesman to our door in the shortest time? No, but generic forms of the TSP have been used in designing circuit boards, reducing waste of raw materials (i.e. lumber or sheet metal) in manufacturing, analyzing crystal structures, scheduling, and much more. An efficient general solution has not been found. The TSP project is undertaking the arduous task of using the brute force method to find an optimal solution to a 48 city TSP (ie, trying all possible routes one at a time). Once the optimal path(s) is/are known, evaluation of other algorithms can begin. The final step for the TSP will be to determine if a combination of algorithms can produce results more quickly.
Image:Enter arrow.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
| uFluids
|
|
Image:Px.gif
|
|
Image:Px.gif
|
Fluid surface tension is typically a very small and ignorable force for most applications. However in space, these forces dominate and cause some very interesting behavior -- look at any NASA film. Also these forces dominate at very small scales and can cause problems with liquid flow through very small (less than 1mm) channels. uFluids is a project to model fluid behavior in the above two cases (low-gravity and at very small scales). Our goals are (1) to facilitate design of better satellite and spacecraft propellant management devices and (2) to optimize performance in microchannel and MEMS (microelectrical-mechanical systems) devices.
Image:Enter arrow.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
| WEP-M+2
|
|
Image:Px.gif
|
|
Image:Px.gif
|
WEP-M+2 (wanless2) is a research project that uses Internet-connected computers to do research in number theory. The project is currently investigating factorization of Mersenneplustwo numbers. Mersenneplustwo numbers are those integers that are two more than a Mersenne prime. Mersenne primes are of the form 2^p-1. This makes Mersenneplustwo numbers of the form 2^p+1. This project aims to find the factors (ie integer divisors) of Mersenneplustwo numbers.
Image:Enter arrow.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
| World Community Grid
|
|
Image:Px.gif
|
|
Image:Px.gif
|
World Community Grid, sponsored by IBM, is a package of medical research projects. You can choose to join any combination: AfricanClimate@Home: Develops more accurate climate models of specific regions in Africa, where negative impacts of climate change are expected to be felt more acutely. Discovering Dengue Drugs - Together: Identifies promising drug leads to combat the related dengue, hepatitis C, West Nile, and Yellow fever viruses. FightAIDS@Home: Uses computational methods to identify compounds that have characteristics necessary to block HIV protease, and thus potential for drug development. Help Conquer Cancer: Improves the results of protein X-ray crystallography, which helps researchers not only annotate unknown parts of the human proteome, but importantly improves their understanding of cancer initiation, progression and treatment. Human Proteome Folding 2: Explores the limits of protein structure prediction, aiming to advance our understanding of these basic building blocks of life.
Image:Enter arrow.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
| XtremLab
|
|
Image:Px.gif
|
|
Image:Px.gif
|
XtremLab is a project which measures the free resources available on personal computers involved in large-scale distributed computing. Ultimately, these measurements will be used to improve the design and implementation of current systems. The project has been able to monitor 32,000 nodes in 18 months. From this experiment we have collected and analyzed more than 17 millions results. NOTE: The project is currently stopped and not collecting new traces. We may resume the project with a new version of the application in the future. Collected data is publicly accessible here.
Image:Enter arrow.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
| Yoyo@home
|
|
Image:Px.gif
|
|
Image:Px.gif
|
Yoyo@home makes available a number of distributed computing projects which would otherwise be incompatible with BOINC and GridRepublic: Muon: Use your PCs idle time to simulate and design parts of a particle accelerator. Evolution@home: Join the first global computing system for evolutionary biology. It allows everybody with an Internet-connected PC to contribute to evolutionary-research by running simulations of evolution. Distributed.net: This project searches for the shortest Optimal Golumb Ruler of the length 25. Note: there may be issues with reporting of statistics from these projects. Note: you may or may not be able to login to the project websites directly. Note: Some virus scanners falsely detect downloads for the Distributed.net project as a "trojan" virus.
Image:Enter arrow.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
|
Image:Px.gif
|
| Zivis
|
|
Image:Px.gif
|
|
Image:Px.gif
|
Zivis is the first "city-wide supercomputer". The project is run by the Zaragoza City Council, and the Institute for Biocomputation and Physics of Complex System (BIFI) at the University of Zaragoza. The objective is to harness local (and non-local) computing resources for local research; and at the same time to involve the community in the science being done locally. The initial research being done on Zivis is on the subject of fusion plasma ("Integration of Stochastic Differential Equations in Plasmas") -- improved understanding of this could lead to better designs for fusion power stations. (Fusion power is a form of nuclear energy that produces a lower volume of less dangerous waste than traditional nuclear fission power.)
Image:Enter arrow.gif
|
|